COVID-19 outbreak in an enclosed environment
Background
An FPSO (Floating production storage and offloading) vessel is a floating production system that is perfectly “enclosed” but needs regular exchange of staffs. It has strict operation schedules and regulations, and has to stop production if it becomes understaffed due to an outbreak of COVID-19.
Assumptions
- The FPSO has 64 staff, with one carrier of the SARS-CoV-2 virus.
- The carrier was recently introduced and did not show any symptom upon arrival.
- Anyone who shows symptom will be removed from the FPSO.
We are interested in knowning:
- How likely that an outbreak will happen.
- Is it enough to remove any symptomatic case.
- Statistics such as how likely a second case will happen after the removal of the first.
Simulation of outbreaks
This scenario can be simulated with the following command:
outbreak_simulator --popsize 64 --infectors 0 --leadtime asymptomatic --repeats 10000 --handle-symptomatic remove
where
--popsizeis the population size--infectoris the individual with the virus, 0 is its ID.--leadtime asymptomaticmeans the individual could be in any stage of infection, as long as he or she does not show any symptom.--handle-symptomatic removeremoves individuals as soon as he or she shows symptom.
We use default options for other key parameters, which includes
--sympatomatic-r0: production number (average number of individuals an infected individual will infect) from1.4to2.8--asymptomatic-r0: production number of asymptomatic cases from0.28 to0.56`.--prop-asym-carriers: proportion of asymptomatic carriers in a simulation, range from0.2to0.4
outbreak_simulator --popsize 64 --infectors 0 --leadtime asymptomatic --repeats 10000 \
--handle-symptomatic remove --logfile enclosed.log > enclosedFPSO_base.txt
100%|████████████████████████████████████| 10000/10000 [00:15<00:00, 634.06it/s]
Results
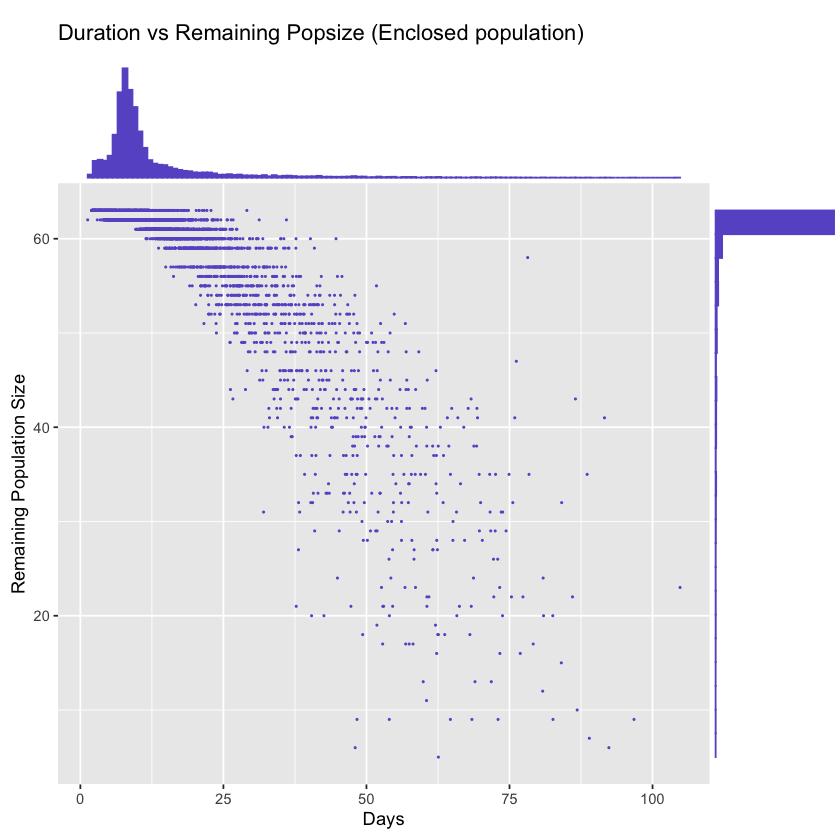
Distribution of duration of outbreak vs remaining population size
Just to show the overall results of simulations, the following figures display the duration of outbreak (x-axis) vs the remaining population size (y-axis) for a few scenarios.
library(ggplot2)
library(ggExtra)
plot_time_vs_size <- function(data_file, title) {
data = read.table(data_file, sep='\t', header=1)
end_count = data[data['event'] == 'END',]
end_count$target = as.numeric(end_count$target)
title = paste0('\nDuration vs Remaining Popsize (', title, ')')
p <- ggplot(end_count, aes(x=time, y=target)) + geom_point(size=0.2, color='slateblue') + theme(legend.position="none") +
xlab('Days') + ylab('Remaining Population Size') + ggtitle(title)
p1 <- ggMarginal(p, type="histogram",
xparams=list(bins=120),
yparams=list(bins=24),
fill = "slateblue",
color="slateblue"
)
print(p1)
}
For example, the the first version of the simulation script, there is a clear lognormal shaped distribution for duration of outbreaks, which indicates the first outbreak will happen around 5 days after the introduction of the seed carrier, and there are likely second and third outbreaks.
plot_time_vs_size('enclosed.log', 'Enclosed population')
Probability of outbreak
The output from the simulator has a large number of summary statistics.
Probability of outbreak
Whether or not an outbreak has happened depend on the definition of outbreak. If we define no outbreak as no one has shown symptom (and not removed),
2116 out of 10000 simulations have a remaining population size of 64. Therefore the probability that an
outbreak will happen is 0.79, which should correspond to the proportion of simulations
with the introduction of asymptomatic cases (30%), and only a small proportion of these cases have infected others and caused an outbreak.
If we define no outbreak as no symptomatic case, or no more cases (and everything returnes to normal) after the removal of the first symptomatic case, 5757 out of 10000 simulations have a remaining population size of 63. Therefore the probability that an outbreak will happen and spread to others is 0.21.
Is removal of symptomatic case enough to stop the outbreak?
We have learned that in 5757 out of 10000 simulations removal of the first symptomatic case actually stopped the spread of the virus (or the virus has spread but no more symptomatic cases would happen). However, among all simulations, despite prompt removal of symptomatic cases, 1536 has a second symptomatic case, and 1187 have a third symptomatic case. That is to say, in 67.7% cases, there would be a second case after the removal of the first symptomatic one, so removal of symptomatic case is unlikely to stop an outbreak.
Availability
This notebook is available under the Applications directory of the GitHub repository of the COVID19 Outbreak Simulator. It can be executed with sos-papermill with the following parameters, or using a docker image bcmictr/outbreak-simulator-notebook as described in here.